The evolution of maternal care and communication
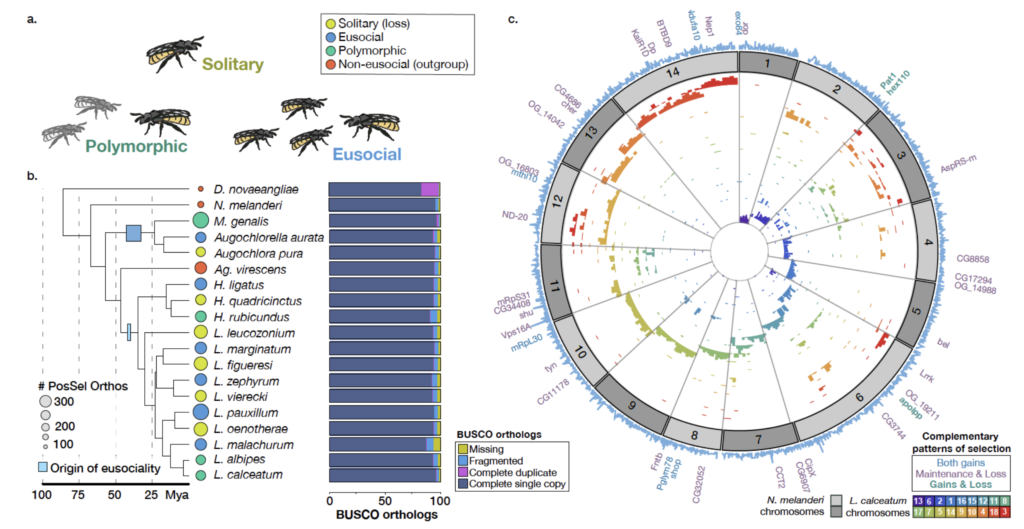
Eusociality, with overlapping generations and a non-reproducing worker caste, represents an extreme form of social behavior that extremely successful, but rare and difficult to evolve. In contrast, maternal care is much more common throughout the animal kingdom, and is thought to be an important precursor to the evolution of eusociality.
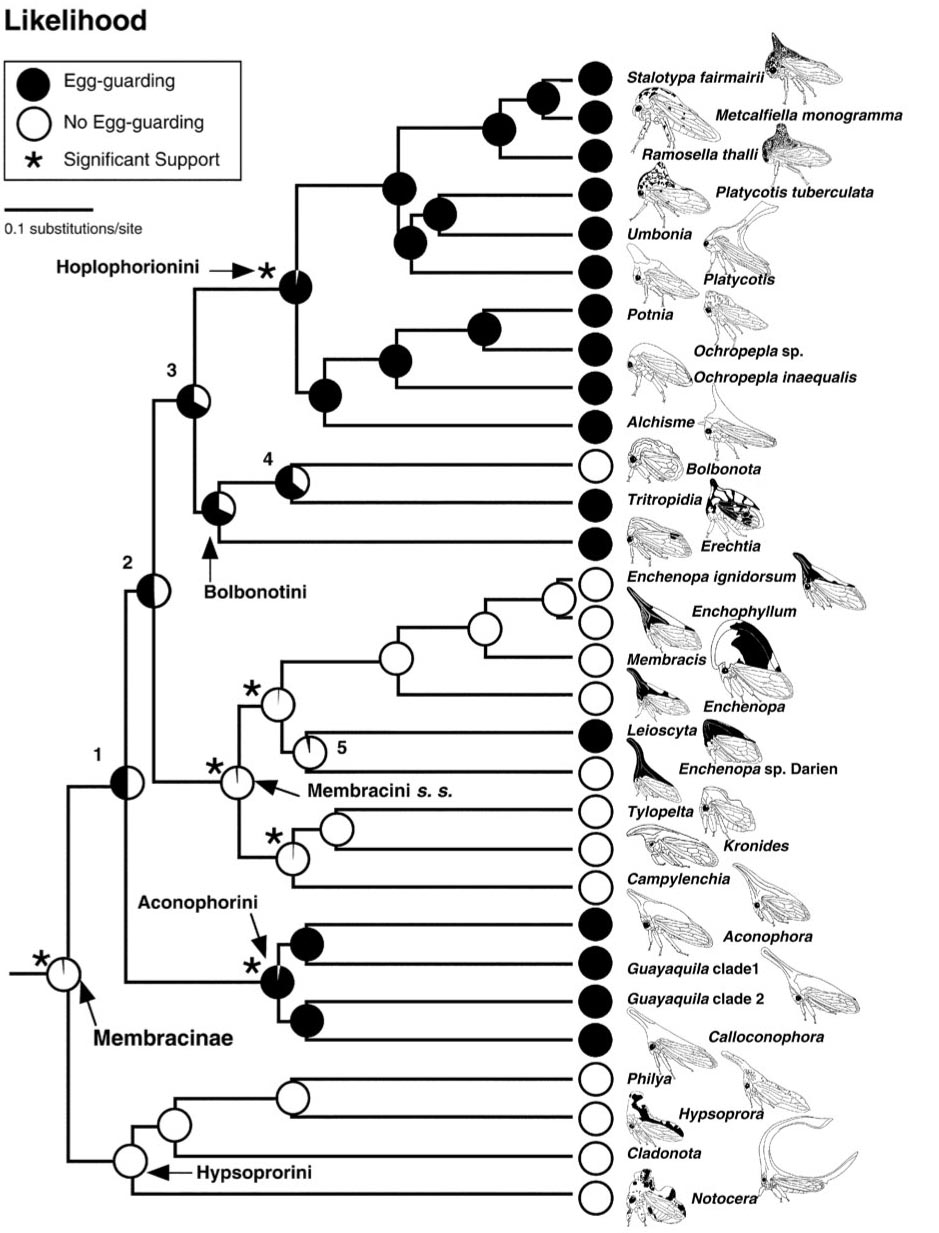
Many insects exhibit some form of maternal care, whether it be egg-guarding or extended provisioning and defense of their offspring. In the treehoppers, maternal care has originated at least 3 times, and like halictids, there have also been many subsequent reversals. This variation creates statistical independence for comparative studies.
Treehoppers exhibit extreme diversity in a number of additional morphological, behavioral, and ecological traits. And like halictid bees, there is also extensive variation in some of these traits within species, opening the door to quantitative and population genetic approaches.
In collaboration with Rex Cocroft (U Missouri, Columbia), we have generated a panel of inbred lines for Tylopelta gibbera, a species that varies in nymphal social behavior and communication. We are using these lines to conduct quantitative genetic and functional studies aimed at identifying the genetic and physiological mechanisms underlying variation in these traits.